Non-negative Matrix Factorisation (NMF) is an unsupervised iterative algorithm that factorises an enter matrix into two matrices with constructive entries. As in PCA, we’re nonetheless deriving new options from our current ones; nevertheless, in PCA, we have been creating orthogonal options primarily based on maximising the defined variance, in NMF, we’re creating non-negative elements and coefficients, that when multiplied give an approximation to the unique information.
Suppose we’ve got an N x M matrix (dataset), to use NMF to this matrix, we select a rank r, and after we apply the algorithm, we’re returned two matrices — the premise matrix W (measurement N x r), and the coefficient matrix H (measurement r x M). Mathematically: A ~ W * H.
The idea matrix, W has measurement N x r, with every of the r columns defining a meta-feature. The coefficient matrix, H has measurement r x M, with every of the M columns representing the meta-feature values of the corresponding samples, that is what we take with us because the lowered dataset. For any rank r, NMF teams the samples into clusters. An vital query that one ought to deal with is whether or not or not a given rank can decompose the samples into significant clusters; this query refers to picking the very best rank to make use of within the algorithm on your particular dataset.
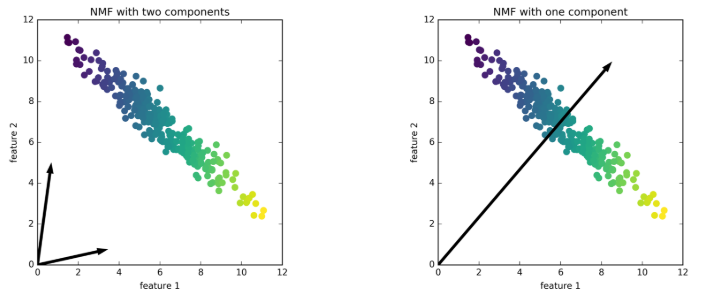
In determine 1, we will see that if we use 2 elements for the artificial dataset, we will simply signify any datapoint, as they may all be linear mixtures of the elements. If the info may be represented in 2 elements, then NMF will create a pair of foundation vectors (elements) that time to the extremes of the info (left image in determine 1). If we solely use one part, then NMF will create a vector (part) that factors to the imply of the info, as this greatest represents the info (proper image in determine 1). Observe that, in contrast to PCA, we should not have a 1st NMF part, the elements should not ordered [1].
The rank is a important parameter. It defines the variety of meta-features used to approximate the dataset. A typical manner of selecting r is to compute some high quality measure of the outcomes and select the very best worth, in line with this measure. Some widespread measures that you need to use can be reconstruction error — compute W * H and calculate the error between the consequence and the unique matrix; one may graph the reconstruction error for a number of ranks and search for an elbow, as in PCA with defined variance ratio. One other measure is Bayesian Info Criterion (BIC) — BIC makes use of the variety of samples, parameters, and the probability of the info to steadiness the goodness of match (measured with the probability) with the complexity of the mannequin — extra complicated fashions are penalised, i.e. fashions with extra parameters. A decrease BIC means a greater mannequin. Additionally, [2] proposed to take the primary worth of r for which the cophenetic correlation begins reducing.
Let’s apply this to a medical dataset with 17 options and 220 samples. We’ll provoke random_state=42 in sklearn.decomposition.NMF(). We run the algorithm on the dataset for a variety of ranks from 2 – 9. We will now compute the metrics and visualise them (see fig. 2).
By observing the graphs, we conclude that r = 4 is the very best rank for our NMF dimension discount. We see a transparent elbow at r = 4 for the reconstruction error; for the BIC curve, we see it takes the worldwide minimal at r = 4; within the cophenetic coefficient curve we see that the graph begins to say no sharply at r = 4. Therefore, we conclude that r = 4 is the very best rank for this dataset. Discover that the three graphs have very completely different patterns and don’t appear like one another, however we will nonetheless use the knowledge supplied by all three to attract a unanimous conclusion — it’s good to attract on a various vary of data to make choices.
There are some apparent drawbacks to this method. We’re restricted to solely non-negative matrices as inputs, this instantly limits the mannequin virtually. Interpretability can be a difficulty; as with PCA, it’s not all the time clear what the elements ‘imply’, particularly if we will’t visualise any outcomes.
NMF may be very helpful for some sorts of information, like pixel information from pictures — as all of the values are non-negative; additionally, NMF may be very helpful for decomposing audio information, akin to an audio clip of a number of individuals talking directly, or a music monitor, as these are examples of information that’s created because the overlay of a number of impartial sources. NMF may give us an approximation of the impartial sources, which may be very helpful in sure settings.
[1] Müller, Andreas C., and Sarah Guido. Introduction to Machine Studying with Python: A Information for Information Scientists. First version. Sebastopol, CA: O’Reilly Media, 2017.
[2] Metagenes and molecular sample discovery utilizing matrix factorisation; Jean-Philippe Brunet, Pablo Tamayo, Todd R. Golub, and Jill P. Mesirov; Massachusetts Institute of Expertise, Cambridge, MA, December 20, 2003.