Within the previous blog, we delved into the foundational ideas of joint chance, conditional chance, and independence, setting the stage for understanding Bayesian Networks.These networks leverage the rules of chance idea to allow sturdy decision-making and inferencing, that are important in numerous machine studying functions.
A Bayesian Community is a graphical mannequin representing a set of variables and their conditional dependencies via a Directed Acyclic Graph (DAG). Every node within the graph corresponds to a variable, and every edge signifies a conditional dependency. The energy of those dependencies is quantified by Conditional Chance Tables (CPTs).
On this second a part of our exploration, we are going to transition from idea to apply by setting up a Bayesian Community step-by-step.
By the top of this information, you should have a transparent understanding of find out how to construct and make the most of Bayesian Networks in sensible eventualities, notably throughout the context of machine studying. Let’s dive in!
Let’s think about an instance of a easy Bayesian community utilizing variables A, B, and C:
we’ll assume the next relationships:
- A influences B.
- A influences C.
- B influences C.
Rationalization
- A: That is the mum or dad node. It influences each B and C.
- B: This node is influenced by A and influences C.
- C: This node is influenced by each A and B.
Conditional Chance Tables (CPTs)
- P(A): The chance of A.
- P(B|A): The chance of B given A.
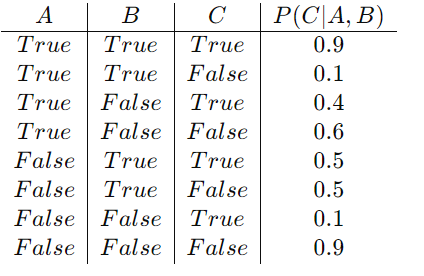
- P(C|A, B): The chance of C given each A and B.
Instance CPTs
Let’s assume binary variables (True/False) for simplicity:
P(A):
P(B|A):
P(C|A, B):
Sensible Utilization
In a sensible state of affairs, you’d use these CPTs to compute the possibilities of various occasions. For instance, to search out the chance that C is true on condition that A is true and B is fake, you’d use the values from the CPT of C:
P(C=True∣A=True,B=False)=0.4
Now , Let’s think about a real-world instance of a probabilistic mannequin utilizing a Bayesian Community to diagnose a medical situation based mostly on signs and check outcomes.We wish to diagnose whether or not a affected person has a illness (D) based mostly on two signs: fever (F) and cough (C), and a check end result (T). The Bayesian Community for this state of affairs might be structured as follows:
- D (Illness) influences F (Fever)
- D (Illness) influences C (Cough)
- D (Illness) influences T (Take a look at end result)
1. Figuring out Variables
- D: Illness (True/False)
- F: Fever (True/False)
- C: Cough (True/False)
- T: Take a look at end result (Constructive/Unfavorable)
2. Figuring out Conditional Dependencies
- P(F∣D): Chance of Fever given Illness
- P(C∣D): Chance of Cough given Illness
- P(T∣D)): Chance of Take a look at end result given Illness
3. Creating the DAG
The Directed Acyclic Graph (DAG) visually represents the dependencies:
4. Defining the CPTs:
Now, I want to present find out how to do it with python.
import torch# Possibilities for D
P_D = torch.tensor([0.01, 0.99]) # P(D=True), P(D=False)
# Conditional chances for F given D
P_F_given_D = torch.tensor([[0.8, 0.2], # P(F=True | D=True), P(F=False | D=True)
[0.1, 0.9]]) # P(F=True | D=False), P(F=False | D=False)
# Conditional chances for C given D
P_C_given_D = torch.tensor([[0.7, 0.3], # P(C=True | D=True), P(C=False | D=True)
[0.2, 0.8]]) # P(C=True | D=False), P(C=False | D=False)
# Conditional chances for T given D
P_T_given_D = torch.tensor([[0.9, 0.1], # P(T=Constructive | D=True), P(T=Unfavorable | D=True)
[0.2, 0.8]]) # P(T=Constructive | D=False), P(T=Unfavorable | D=False)
Now, we’ll outline capabilities to pattern from the distributions and compute joint chances.
def sample_D():
return torch.distributions.Bernoulli(P_D[0]).pattern().merchandise()def sample_F(D):
return torch.distributions.Bernoulli(P_F_given_D[D][0]).pattern().merchandise()
def sample_C(D):
return torch.distributions.Bernoulli(P_C_given_D[D][0]).pattern().merchandise()
def sample_T(D):
return torch.distributions.Bernoulli(P_T_given_D[D][0]).pattern().merchandise()
def joint_probability(D, F, C, T):
p_D = P_D[D]
p_F_given_D = P_F_given_D[D, F]
p_C_given_D = P_C_given_D[D, C]
p_T_given_D = P_T_given_D[D, T]
return p_D * p_F_given_D * p_C_given_D * p_T_given_D
We’ll pattern from the community and compute the joint chance for a given set of variables.
# Pattern from the community
D = int(sample_D())
F = int(sample_F(D))
C = int(sample_C(D))
T = int(sample_T(D))# Compute joint chance P(D, F, C, T)
jp = joint_probability(D, F, C, T)
print(f"Sampled D: {D}")
print(f"Sampled F: {F}")
print(f"Sampled C: {C}")
print(f"Sampled T: {T}")
print(f"Joint Chance P(D, F, C, T): {jp.merchandise()}")
output:
Sampled D: 0
Sampled F: 1
Sampled C: 0
Sampled T: 0
Joint Chance P(D, F, C, T): 0.0012600000482052565
The sampled values counsel that the affected person doesn’t exhibit a cough (C = False) and obtained a unfavourable check end result (T = Unfavorable). Regardless of experiencing fever (F = True), the chance of the affected person having the illness (D) is low (P(D = False, F = True, C = False, T = Unfavorable) = 0.00126). This inference signifies that whereas fever could also be current, the absence of cough and a unfavourable check end result strongly mitigate the probability of the illness being current on this explicit case. These insights exhibit the utility of Bayesian Networks in probabilistic reasoning, permitting us to interpret complicated relationships between signs, check outcomes, and illness presence successfully.
Bayesian networks present a structured approach to characterize and cause in regards to the conditional dependencies between variables. Understanding the relationships and conditional chances between variables helps in constructing sturdy probabilistic fashions for numerous functions in machine studying and past.