If you’re in information science, there are excessive possibilities of utilizing pandas in your Information Science and Machine Studying processes and information pipelines. Contemplating the necessity to consult with syntax and the fundamentals of pandas, here’s a fast 10-minute intro to pandas and their most used strategies.
Observe: On this article, “pd” is an alias for pandas and “np” is an alias for Numpy.
Making a Sequence by passing an inventory of values, letting pandas create a default integer index:
sequence = pd.Sequence([1,3,5,np.nan,6,8])
sequence
0 1.0
1 3.0
2 5.0
3 NaN
4 6.0
5 8.0
dtype: float64dates = pd.date_range('20130101', intervals=6)
datesDatetimeIndex(['2013-01-01', '2013-01-02', '2013-01-03', '2013-01-04',
'2013-01-05', '2013-01-06'],
dtype='datetime64[ns]', freq='D') test_df = pd.DataFrame(np.random.randn(6,4), index=dates, columns=checklist('ABCD'))
test_dfDate A B C D
2013-01-01 -0.165045 0.286237 -0.388395 0.189089
2013-01-02 -0.380108 0.781734 -0.668664 0.122847
2013-01-03 1.982129 1.970573 1.724951 -0.810865
2013-01-04 -1.390268 -0.862023 1.708512 -1.268239
2013-01-05 1.007223 0.024108 0.539417 1.442396
2013-01-06 1.223380 -0.034152 0.349011 -0.225668
Right here is methods to view the highest and backside rows of the body.
df.head()
df.tail(3)
Date A B C D
2013-01-03 1.982129 1.970573 1.724951 -0.810865
2013-01-04 -1.390268 -0.862023 1.708512 -1.268239
2013-01-05 1.007223 0.024108 0.539417 1.442396
Show the index, columns, and the underlying NumPy information:
df.index
DatetimeIndex(['2013-01-01', '2013-01-02', '2013-01-03', '2013-01-04',
'2013-01-05', '2013-01-06'],
dtype='datetime64[ns]', freq='D')df.columnsIndex(['A', 'B', 'C', 'D'], dtype='object')df.valuesarray([[-0.16504516, 0.28623677, -0.38839496, 0.1890891 ],
[-0.38010769, 0.78173448, -0.66866431, 0.12284665],
[ 1.98212925, 1.9705729 , 1.72495074, -0.81086545],
[-1.39026802, -0.86202321, 1.70851228, -1.26823932],
[ 1.0072233 , 0.02410772, 0.53941737, 1.44239551],
[ 1.22337986, -0.03415161, 0.34901142, -0.22566768]])
describe() exhibits a fast statistical abstract of your information.
df.describe()
A B C D
rely 6.000000 6.000000 6.000000 6.000000
imply 0.379552 0.361080 0.544139 -0.091740
std 1.239371 0.952760 1.012787 0.937839
min -1.390268 -0.862023 -0.668664 -1.268239
25% -0.326342 -0.019587 -0.204043 -0.664566
50% 0.421089 0.155172 0.444214 -0.051411
75% 1.169341 0.657860 1.416239 0.172528
max 1.982129 1.970573 1.724951 1.442396
Transposing your information;
df.T
Sorting by an axis;
df.sort_index(axis=1, ascending=False)
Date A B C D
2013-01-01 0.189089 -0.388395 0.286237 -0.165045
2013-01-02 0.122847 -0.668664 0.781734 -0.380108
2013-01-03 -0.810865 1.724951 1.970573 1.982129
2013-01-04 -1.268239 1.708512 -0.862023 -1.390268
2013-01-05 1.442396 0.539417 0.024108 1.007223
2013-01-06 -0.225668 0.349011 -0.034152 1.223380
Sorting by values;
df.sort_values(by='B')
Date A B C D
2013-01-04 -1.390268 -0.862023 1.708512 -1.268239
2013-01-06 1.223380 -0.034152 0.349011 -0.225668
2013-01-05 1.007223 0.024108 0.539417 1.442396
2013-01-01 -0.165045 0.286237 -0.388395 0.189089
2013-01-02 -0.380108 0.781734 -0.668664 0.122847
2013-01-03 1.982129 1.970573 1.724951 -0.810865
Whereas Customary Python / Numpy expressions for choosing and setting are intuitive and come in useful for interactive work, for manufacturing code, it is strongly recommended to make use of the optimized Pandas information entry strategies equivalent to .at, .iat, .loc, and many others…
Choosing a single column, which yields a Sequence, equal to df. A.
df['A']
2013-01-01 -0.165045
2013-01-02 -0.380108
2013-01-03 1.982129
2013-01-04 -1.390268
2013-01-05 1.007223
2013-01-06 1.223380
Freq: D, Identify: A, dtype: float64
Choosing by way of [], which slices the rows.
df[0:3]
Date A B C D
2013-01-01 -0.165045 0.286237 -0.388395 0.189089
2013-01-02 0.380108 0.781734 -0.668664 0.122847
2013-01-03 1.982129 1.970573 1.724951 -0.810865
Choice by Label for getting a cross-section utilizing a label;
df.loc[dates[0]]
A -0.165045
B 0.286237
C -0.388395
D 0.189089
Identify: 2013-01-01 00:00:00, dtype: float64
Choosing on a multi-axis by label:
df.loc[:,['A','B']]
Date A B
2013-01-01 -0.165045 0.286237
2013-01-02 -0.380108 0.781734
2013-01-03 1.982129 1.970573
2013-01-04 -1.390268 -0.862023
2013-01-05 1.007223 0.024108
2013-01-06 1.223380 -0.034152
Displaying label slicing, each endpoints are included:
df.loc['20130102':'20130104',['A','B']]
Date A B
2013-01-02 -0.380108 0.781734
2013-01-03 1.982129 1.970573
2013-01-04 -1.390268 -0.862023
Discount within the dimensions of the returned object.
df.loc['20130102',['A','B']]
A -0.380108
B 0.781734
Identify: 2013-01-02 00:00:00, dtype: float64
For getting a scalar worth:
df.loc[dates[0],'A']
For getting quick entry to a scalar (equal to the prior methodology):
df.at[dates[0],'A']
Choose by way of the place of the handed integers:
df.iloc[3]
A -1.390268
B -0.862023
C 1.708512
D -1.268239
Identify: 2013-01-04 00:00:00, dtype: float64
By integer slices, appearing just like numpy/python:
df.iloc[3:5,0:2]
Date A B
2013-01-04 -1.390268 -0.862023
2013-01-05 1.007223 0.024108
By lists of integer place places, just like the numpy/python type:
df.iloc[[1,2,4],[0,2]]
Date A C
2013-01-02 -0.380108 -0.668664
2013-01-03 1.982129 1.724951
2013-01-05 1.007223 0.539417
For slicing rows explicitly:
df.iloc[1:3,:]
Date A B C D
2013-01-02 -0.380108 0.781734 -0.668664 0.122847
2013-01-03 1.982129 1.970573 1.724951 -0.810865
For slicing columns explicitly:
df.iloc[:,1:3]
Date A B
2013-01-01 0.286237 -0.388395
2013-01-02 0.781734 - 0.668664
2013-01-03 1.970573 1.724951
2013-01-04 -0.862023 1.708512
2013-01-05 0.024108 0.539417
2013-01-06 -0.034152 0.349011
Utilizing a single column’s values to pick out information.
df[df.A > 0]
Date A B C D
2013-01-03 1.982129 1.970573 1.724951 -0.810865
2013-01-05 1.007223 0.024108 0.539417 1.442396
2013-01-06 1.223380 -0.034152 0.349011 -0.225668
Choosing values from a DataFrame the place a Boolean situation is met.
df[df > 0]
Date A B C D
2013-01-01 NaN 0.286237 NaN 0.189089
2013-01-02 NaN 0.781734 NaN 0.122847
2013-01-03 1.982129 1.970573 1.724951 NaN
2013-01-04 NaN NaN 1.708512 NaN
2013-01-05 1.007223 0.024108 0.539417 1.442396
2013-01-06 1.223380 NaN 0.349011 NaN
Utilizing the isin() methodology for filtering:
df2 = df.copy()
df2['E'] = ['one', 'one','two','three','four','three']
df2
Date A B C D E
2013-01-01 -0.165045 0.286237 -0.388395 0.189089 one
2013-01-02 -0.380108 0.781734 -0.668664 0.122847 one
2013-01-03 1.982129 1.970573 1.724951 -0.810865 two
2013-01-04 -1.390268 -0.862023 1.708512 -1.268239 three
2013-01-05 1.007223 0.024108 0.539417 1.442396 4
2013-01-06 1.223380 -0.034152 0.349011 -0.225668 three
df2[df2['E'].isin(['two','four'])]
Date A B C D E
2013-01-03 1.982129 1.970573 1.724951 -0.810865 two
2013-01-05 1.007223 0.024108 0.539417 1.442396 4
Setting a brand new column routinely aligns the info with the indexes.
new_series = pd.Sequence([1,2,3,4,5,6], index=pd.date_range('20130102', intervals=6))
new_series
2013-01-02 1
2013-01-03 2
2013-01-04 3
2013-01-05 4
2013-01-06 5
2013-01-07 6
Freq: D, dtype: int64
Setting values by label:
df.at[dates[0],'A'] = 0
Setting by assigning with a NumPy array:
df.loc[:,'D'] = np.array([5] * len(df))
Pandas primarily use the worth np.nan to symbolize lacking information. It’s by default not included in computations. Reindexing permits you to change/add/delete the index on a specified axis. This returns a duplicate of the info.
df = df.reindex(index=dates[0:4], columns=checklist(df.columns) + ['E'])
df.loc[dates[0]:dates[1],'E'] = 1
df
Date A B C D E F
2013-01-01 0.000000 0.000000 -0.388395 5 NaN 1.0
2013-01-02 -0.380108 0.781734 -0.668664 5 1.0 1.0
2013-01-03 1.982129 1.970573 1.724951 5 2.0 NaN
2013-01-04 -1.390268 -0.862023 1.708512 5 3.0 NaN
To drop any rows which have lacking information.
df.dropna(how='any')
Date A B C D E F
2013-01-02 -0.380108 0.781734 -0.668664 5 1.0 1.0
Filling lacking information.
df.fillna(worth=5)
Date A B C D E F
2013-01-01 0.000000 0.000000 -0.388395 5 5.0 1.0
2013-01-02 -0.380108 0.781734 -0.668664 5 1.0 1.0
2013-01-03 1.982129 1.970573 1.724951 5 2.0 5.0
2013-01-04 -1.390268 -0.862023 1.708512 5 3.0 5.0
To get the boolean masks the place values are nan.
pd.isna(df)
Date A B C D E F
2013-01-01 False False False False True False
2013-01-02 False False False False False False
2013-01-03 False False False False False True
2013-01-04 False False False False False True
Making use of capabilities to the info:
df.apply(np.cumsum)
Date A B C D E
2013-01-01 0.000000 0.000000 -0.388395 5 NaN
2013-01-02 -0.380108 0.781734 -1.057059 10 1.0
2013-01-03 1.602022 2.752307 0.667891 15 3.0
df.apply(lambda x: x.max() - x.min())
Sequence is supplied with a set of string processing strategies within the str attribute that make it simple to function on every aspect of the array. Observe that pattern-matching in str usually makes use of common expressions by default.
str_series = pd.Sequence(['A', 'B', 'C', 'Aaba', 'Baca', np.nan, 'CABA', 'dog', 'cat'])
str_series0 A
1 B
2 C
3 Aaba
4 Baca
5 NaN
6 CABA
7 canine
8 cat
dtype: objects.str.decrease()0 a
1 b
2 c
3 aaba
4 baca
5 NaN
6 caba
7 canine
8 cat
dtype: object
Pandas present numerous amenities for simply combining collectively Sequence, DataFrame, and Panel objects with numerous sorts of set logic for the indexes and relational algebra performance within the case of be part of / merge-type operations. Concatenating pandas objects along with concat():
df = pd.DataFrame(np.random.randn(10, 4))
break it into items
items = [df[:3], df[3:7], df[7:]]
pd.concat(items)
A B C D
-0.106234 -0.950631 1.519573 0.097218
1.796956 -0.450472 -1.315292 -1.099288
1.589803 0.774019 0.009430 -0.227336
1.153811 0.272446 1.984570 -0.039846
0.495798 0.714185 -1.035842 0.101935
0.254143 0.359573 -1.274558 -1.978555
0.456850 -0.094249 0.665324 0.226110
-0.657296 0.760446 -0.521526 0.392031
0.186656 -0.131740 -1.404915 0.501818
-0.523582 -0.876016 -0.004513 -0.509841
left = pd.DataFrame({'key': ['foo', 'foo'], 'lval': [1, 2]})
proper = pd.DataFrame({'key': ['foo', 'foo'], 'rval': [4, 5]})
key lval
foo 1
foo 2key rval
foo 4
foo 5
pd.merge(left, proper, on='key')
key lval rval
foo 1 4
foo 1 5
foo 2 4
foo 2 5
Append rows to a dataframe.
append_df = pd.DataFrame(np.random.randn(8, 4), columns=['A','B','C','D'])
append_df
A B C D
0.310213 0.511346 1.891497 0.491886
-2.099571 -0.477107 0.701392 0.452229
-1.508507 0.207553 0.140408 0.033682
-1.026017 -1.277501 1.755467 1.056045
-0.890034 0.726291 -0.419684 -1.073366
-0.614249 1.139664 -1.582946 0.661833
-0.010116 1.877924 -0.015332 1.176713
-0.314318 1.088290 -0.067972 -1.759359
By “group by” we’re referring to a course of involving a number of of the next steps:
- Splitting the info into teams primarily based on some standards.
- Making use of a operate to every group independently.
- Combining the outcomes into a knowledge construction.
df = pd.DataFrame({'A' : ['foo', 'bar', 'foo', 'bar', 'foo', 'bar', 'foo', 'foo'],
'B' : ['one', 'one', 'two', 'three', 'two', 'two', 'one', 'three'],
'C' : np.random.randn(8),
'D' : np.random.randn(8)})
A B C D
foo one -0.606619 0.295979
bar one -0.015111 -1.662742
foo two -0.212922 1.564823
bar three 0.332831 0.337342
foo two 0.235074 -0.568002
bar two -0.892237 0.944328
foo one 0.558490 0.977741
foo three 0.517773 1.052036
Grouping after which making use of the sum() operate to the ensuing teams.
df.groupby('A').sum()
C D
A
bar -0.574517 -0.381072
foo 0.491797 3.322576
Grouping by a number of columns varieties a hierarchical index, and once more we are able to apply the sum operate.
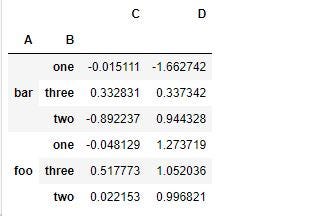
df.groupby(['A','B']).sum()
tuples = checklist(zip(*[['bar', 'bar', 'baz', 'baz', 'foo', 'foo', 'qux', 'qux'],
['one', 'two', 'one', 'two', 'one', 'two', 'one', 'two']]))
tuples[('bar', 'one'),
('bar', 'two'),
('baz', 'one'),
('baz', 'two'),
('foo', 'one'),
('foo', 'two'),
('qux', 'one'),
('qux', 'two')]index = pd.MultiIndex.from_tuples(tuples, names=['first', 'second'])
df = pd.DataFrame(np.random.randn(8, 2), index=index, columns=['A', 'B'])df_ind = df[:4]df_ind
The stack() methodology “compresses” a stage within the DataFrame’s columns.
stacked = df_ind.stack()
stacked
Writing to a CSV file.
df.to_csv('foo.csv')
Studying a CSV
pd.read_csv('foo.csv')
Writing to an HDF5 Retailer.
df.to_hdf('foo.h5','df')
Studying from an Excel file
pd.read_excel('foo.xlsx', 'Sheet1', index_col=None, na_values=['NA'])
Pandas is certainly a robust bundle to work with, particularly for information engineers, and scientists who work on manipulating and analysing information. With a strong grasp of Pandas, you might be well-equipped to streamline your information workflow and uncover beneficial insights out of your information.